Large String Matcher
Python
Dynamic Programming Algorithms
Divide and Conquer Algorithms
Summary
This project was created for my Analysis of Algorithms class at USC Viterbi School of Engineering.
The goal of this project was to implement in code a dynamic programming algorithm that rated the relative similarity between two strings. Use cases for an efficient algorithm would be for matching the similarity between strands of DNA.
The Problem
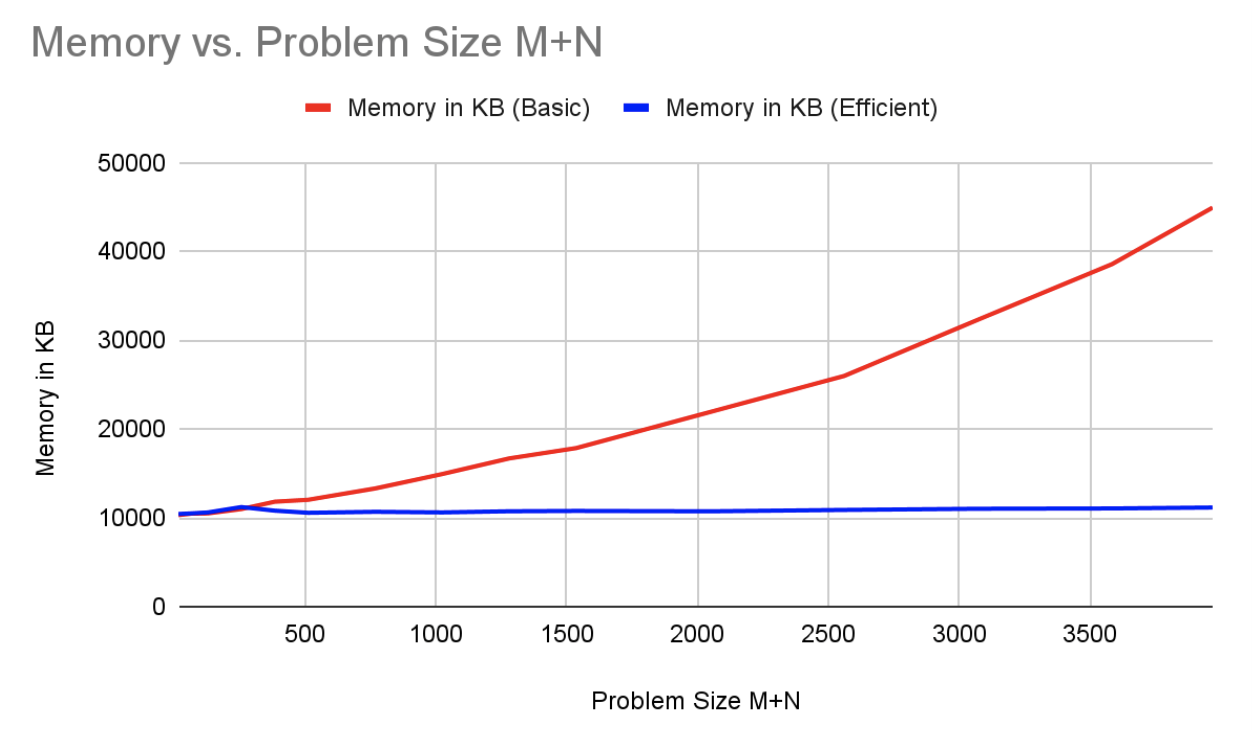
While a basic algorithm is easy to implement, it has its drawbacks. A dynamic programming algorithm uses memoization to record data. The amount of memory required for this algorithm is an array of size M*N where M and N are the lengths of the two input strings. While this is fine for small strings, DNA strands can be millions of strings long, requiring a lot of memory.
Basic Algorithm:
OPT(i, j) = min(OPT(i-1, j-1) + alpha(i, j), OPT(i-1, j) + delta, OPT(i, j-1) + delta)
The Solution
Creating an improved version of the algorithm requires a divide-and-conquer approach. The algorithm splits the first string in half then finds the optimal point to split the second string before finding the optimal alignment between the split sides. This is recursively repeated until one of the two sides has less than two characters. The optimal alignment can then be returned from the recursive base case, resulting in an optimal alignment for the whole string.
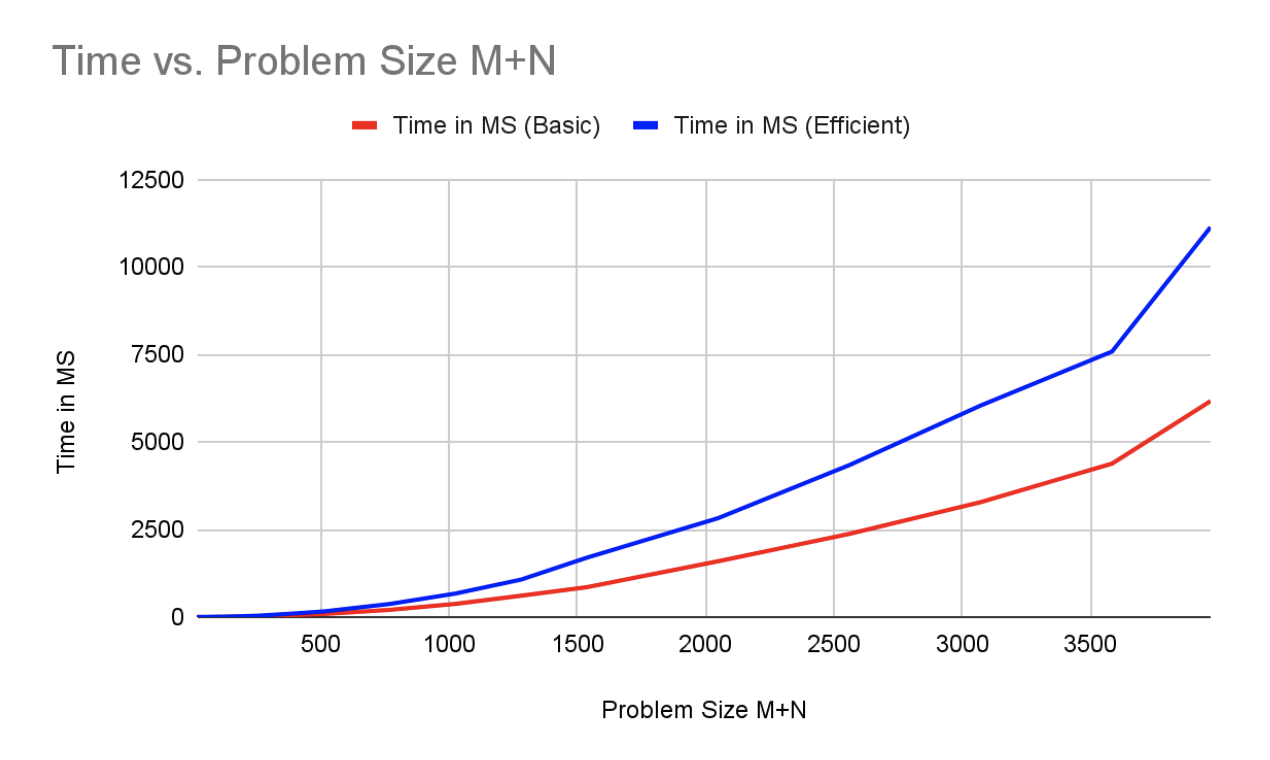
This approach reduces the memory requirement from M*N to 2*N, a significant improvement for very long strings. The tradeoff is the runtime is increased by a polynomial amount.
| M+N | Time in MS (Basic) | Time in MS (Memory Efficient) | Memory in KB (Basic) | Memory in KB (Memory Efficient) |
|---|---|---|---|---|
| 16 | 0.3561973571777344 | 0.3972053527832031 | 10308 | 10436 |
| 64 | 1.683950424194336 | 3.08990478515625 | 10468 | 10500 |
| 128 | 5.637884140014648 | 10.607004165649414 | 10500 | 10608 |
| 256 | 20.433902740478516 | 39.086103439331055 | 10988 | 11216 |
| 384 | 51.07474327087402 | 96.16994857788086 | 11820 | 10808 |
| 512 | 86.45486831665039 | 160.65096855163574 | 12036 | 10568 |
| 768 | 204.65803146362305 | 372.5087642669678 | 13312 | 10680 |
| 1024 | 374.7079372406006 | 670.6089973449707 | 14932 | 10612 |
| 1280 | 609.0717315673828 | 1069.2028999328613 | 16696 | 10740 |
| 1536 | 852.2140979766846 | 1694.1630840301514 | 17856 | 10780 |
| 2048 | 1592.8051471710205 | 2824.464797973633 | 21924 | 10736 |
| 2560 | 2375.648021697998 | 4341.222763061523 | 25980 | 10892 |
| 3072 | 3282.5939655303955 | 6043.540000915527 | 32324 | 11024 |
| 3584 | 4383.219957351685 | 7586.020946502686 | 38576 | 11060 |
| 3968 | 6169.418096542358 | 11138.368844985962 | 44940 | 11164 |